Me vs. the EGA part 3: winning
If you’ve been following these posts so far, you’ll have XML files that register a study and sample samples, and an XML file that registers an analysis (i.e. that lists all the actual data files). And you’ve uploaded all your files to your FTP inbox, and you’ve run them through the test service, and it all responded without any errors.
Right?
Right. So now you need to join all this into datasets.
Anti-aliasing
Since we’ve generated a unique ‘alias’ for each object (studies, samples, analysis files) we should be able use that to link our datasets together, right? Wrong. For the dataset XML we need the accessions.
To get accessions for the analysis files, we need to actually submit all the stuff we had already. I mean you could do that by submitting to the real (non test) service, like this:
$ curl \
-u <ega-box-XXX>:<password> \
-F "SUBMISSION=@submit.xml" \
-F "[THING]=@[XML filename]" \
https://www.ebi.ac.uk/ena/submit/drop-box/submit/
and capturing the response, which is another XML file, and parsing it to get the accessions, but there’s an easier way.
The easier way to submit
Go to this EGA Webin site and log in. You’ll see a ‘Submit’ tag and a ‘Submit XML files’ button. They give us a way to submit the XML files and get useful results back.
Start by choosing your submission xml (submit.xml) and your study xml (study.xml) described in
part one.
Submit them. You get back a tab-separated file giving you the accession for your study, and
also a receipt file. Save them.
Now do the same for each of the ‘samples’ and ‘analysis’ XMLs in turn. (To do this you have to
unselect the XML you’ve already selected. I couldn’t see a way to do this except by Ctrl-R reloading
the page, but that seems to work.) You get back more accessions. Save them.
So now you’ve got files listing the accessions - one for the study, one for each of the samples, and importantly, one for each of the analysis objects (files) in your dataset.
Endgame
This is now pretty straightforward. You basically want this:
<DATASETS>
<DATASET alias="My cool datasets" center_name="<center name>" broker_name="EGA">
<TITLE>My cool dataset</TITLE>
<DATASET_TYPE>Whole genome sequencing</DATASET_TYPE>
<ANALYSIS_REF accession="<accession of first analysis>" />
<ANALYSIS_REF accession="<accession of second analysis>" />
...
<POLICY_REF accession="<accession of data access policy>" refcenter="<center name>"/>
<DATASET_LINKS>
<DATASET_LINK>
<URL_LINK>
<LABEL>My website</LABEL>
<URL><URL of my website></URL>
</URL_LINK>
</DATASET_LINK>
</DATASET_LINKS>
</DATASET>
...
</DATASETS>
Repeat the <DATASET> for as many datasets* as you have.
* Given the presence of <DATASET_TYPE> in there, I was worried I wouldn’t be able to
include all my desired files in the same dataset. This worry seems to have been unfounded - I
included a bunch of analysis specifying CRAM files, and and analysis specifying the VCF of
microarray genotypes and the README file and two annotation files, and it seems to work.
Before writing the XML above you need to know:
- the accession of all your analyses (from the submission step above)
- and the accession of your data access policy. Here I ran into a couple of issues:
Policy issues
First, although there’s an EGA page listing data access commitees, there’s no such page listing policies. So if you’ve got an existing policy but don’t know its accession, you have to ask the EGA helpdesk.
Second, even if you do have a policy, the submission system might not know about it. Advice from the EGA helpdesk was that this happens because these objects are too old, and were entered manually, and the procedure has now changed completely. However this isn’t much help when uploading data.
I’ve chosen to workaround this by creating a new data access committee and a new data access policy. Then I’m going to email the helpdesk and ask them to link it to the correct policy and DAC when the data is processed. Luckily the two XMLs used for this are pretty simple:
<DAC_SET>
<DAC alias="Placeholder for EGAC<<my policy number>" center_name="<center_name>" broker_name="EGA">
<TITLE>My data access committee</TITLE>
<CONTACTS>
<CONTACT name="My IDAC" email="<my dac email address>" organisation="<my organisation>" telephone_number=""/>
</CONTACTS>
</DAC>
</DAC_SET>
and
<POLICY_SET>
<POLICY alias="Placeholder for EGAP<<my policy number>" center_name="<center_name>" broker_name="EGA">
<TITLE>My data access policy</TITLE>
<DAC_REF accession="EGAC00001001201" refcenter="<center_name>"/>
<POLICY_TEXT>See policy EGAP<<my policy number></POLICY_TEXT>
<POLICY_LINKS>
<POLICY_LINK>
<URL_LINK>
<LABEL>Data Access Agreement</LABEL>
<URL><my policy URL></URL>
</URL_LINK>
</POLICY_LINK>
</POLICY_LINKS>
</POLICY>
</POLICY_SET>
(Note that although the dataset only needs the policy, not the DAC, you need to do the DAC because otherwise you can’t creat the policy).
These two XMLs are pretty easy to submit, and now you’ve got a policy accession to include in your dataset.
Return of the work/life balance
Check it out: 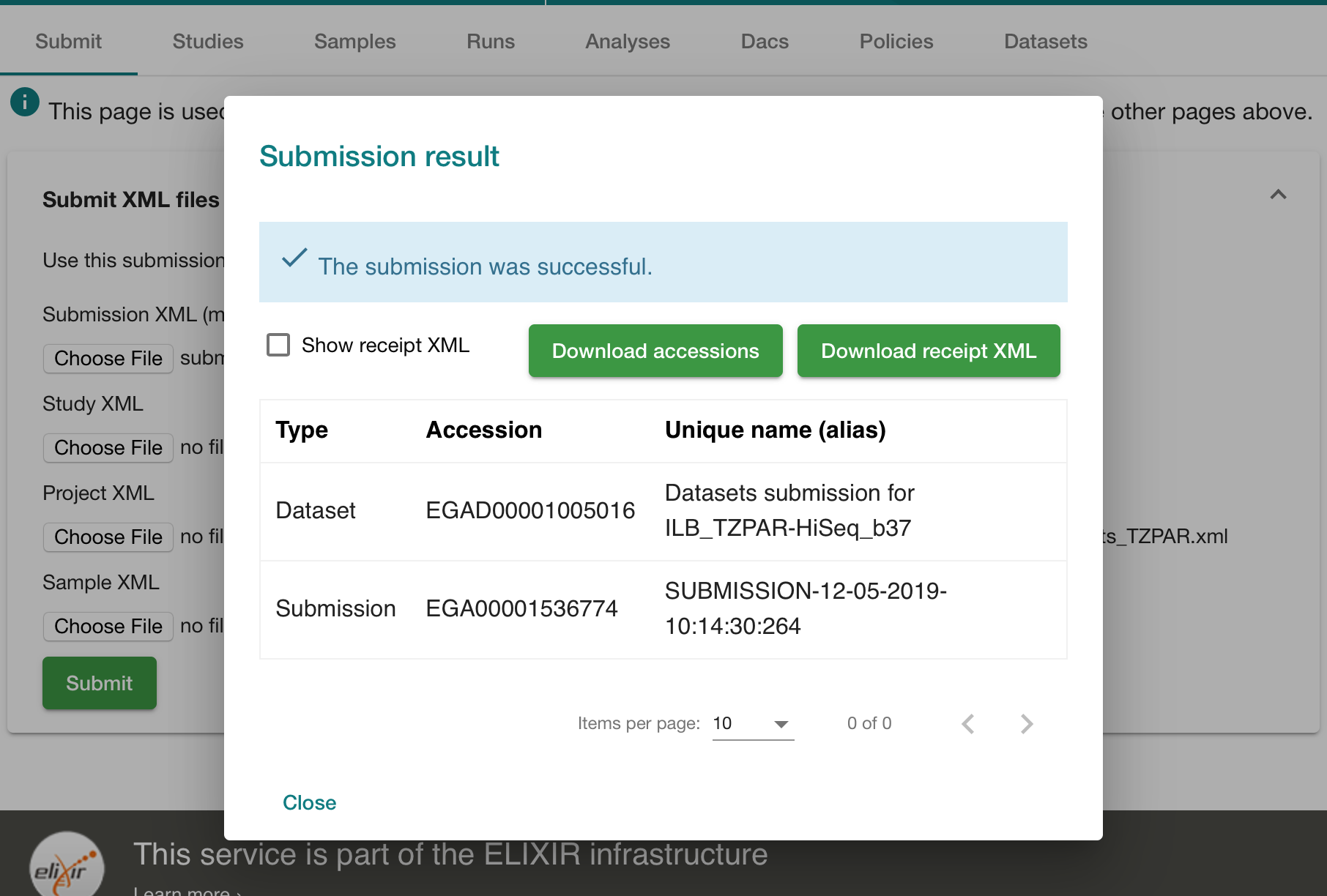
Sit back. Take a deep breath. Now go and play football.
** END **